SCIENCE BRINGS NATIONS TOGETHER
Modern information technologies in biology and medicine
The international workshop “Modern information technologies in biology and medicine” will take place from 22 to 24 November 2023. There are oral and practice parts. During 3 days the specialists from JINR, Serbian and Russian institutes will share their views, outcomes concerning contemporary IT methods, and approaches in application for research. Topics include various scientific fields such as biomedicine, information technology and physics. The workshop is going to be online. The idea is to show the opportunity of application IT solutions for different scientific branches as well as be inspired to search for something new in a friendly ambience. On the 3rd day, the specialists of Meshcheryakov Laboratory of Information Technologies (MLIT) organize the tutorial for members who would like to try the use of Python for machine learning in biomedicine task.
The video conference room
https://jinr.mts-link.ru/2271283/210907340
ORGANIZING COMMITTEE:
Chairman: Oksana Streltsova (MLIT, JINR)
Vice-chairman: Marko Ćosić (Vinča Institute of Nuclear Sciences, Serbia)
Scientific Secretary: Inna Kolesnikova (LRB, JINR)
Nechaevskiy Andrey (MLIT, JINR / Dubna State University)
Podgainy Dmitry (MLIT, JINR)
Priakhina Daria (MLIT, JINR)
Sanja Despotović (Institute for Histology and Embryology, Serbia)
Zuev Maxim (MLIT, JINR)
-
-
10:00
→
10:05
Welcoming address of the director of LRB JINR 5mSpeaker: Aleksandr Bugay (Joint Institute for Nuclear Research)
-
10:05
→
10:10
Welcoming address of the director of Vinča Institute 5mSpeaker: Dr Miroslav Adžić (Vinča Nuclear Research Institute)
-
10:10
→
10:40
Overview of the activities of the Laboratory of Radiation Biology 30m
The presence of a wide range of radiation sources, and above all, beams of heavy ions of various energies at the JINR basic facilities, provides a unique opportunity to solve the fundamental problems of modern radiobiology, astrobiology, neurophysiology, molecular biology and genetics, as well as practical applications in radiation medicine and radiation risk assessments on Earth and in space.
The research program of the Laboratory of Radiation Biology is aimed at studying the mechanisms of action of ionizing radiation with different physical characteristics at the molecular, cellular, tissue and organismal levels of biological organization.
Within the framework of the research program, the following fundamental and applied problems of modern radiation biology will be addressed: studies of formation and repair of clustered DNA damage after exposure of normal and tumor cells of mammals and humans to radiations with different characteristics; studies of radiation-induced gene, structural and complex mutations in mammalian and human cells; studies of violations of behavioral reactions and pathomorphological changes in various brain structures, critical organs and systems of irradiated animals under normal conditions and under the action of radioprotectors; studies of the mechanisms of neurodegeneration under the action of ionizing radiations; development of new approaches to improving the biological effectiveness of radiation therapy of tumors and methods of targeted delivery of radiopharmaceuticals; development of new setups and dosimetry systems for irradiating biological samples, methods for non-destructive analysis of unique samples, and test systems for automated computer processing of biological data; formulation of new mathematical models and computational approaches for radiobiology, bioinformatics and radiation medicine; identification of mechanisms and pathways of catalytic synthesis of prebiotic compounds under the action of radiation.
References
[1] The Laboratory of Radiation Biology. Official web-site http://lrb.jinr.ru/index.php/en/
[2] Brochure “Radiobiological research at JINR” 2023 http://www.jinr.ru/wp-content/uploads/Brochures/JINR_A4_brochure_radiobiology_2023_eng.pdfSpeaker: Aleksandr Bugay (Joint Institute for Nuclear Research) -
10:40
→
11:10
The use of modern information technologies for applied radiobiological research 30m
The study of radiobiological effects on the cell and organismal levels includes various procedures and approaches. The outcomes of such biomedical research are an essential task, primarily associated with computer diagnostics. We face with a need to analyze of a large number experimental data, save and store it as well as to have an opportunity of sharing with the members. The procedure for automatic counting of behavioral patterns and brain cells is a key step in the systems of behavioral analysis of videos and microscopic analysis of medical images of histological slides. In this regard, the main goal of the work is to develop effective methods for the automatic determination and quantification of behavioral patterns as well as brain cells based on modern methods of video/ image pre-processing, processing, computer view and modern approaches of artificial neural network.
One more reason is the difficulty in analyzing experimental heterogeneous data, which includes morphological data (images of sections of various biological tissues), behavioral data (video of experimental animals), etc., obtained by various groups of researchers. A complete understanding of the impact process and a qualitative picture of the effects of ionizing radiation on bio-systems require the systematization and simultaneous processing of a significant amount of these data relating to various aspects of the demonstration of the exposure. The use contemporary IT methods and approaches seem promising. The ordinary radiobiological study includes a set of methods to work with the laboratory rodents, behavioral tests, preparing biological samples, histological analysis, systematizing and analyzing experimental data, presenting data in a form convenient for complex statistical analysis. The outcome of the study is learning the basics of behavioral testing as well as histological technique and analysis, mark-up the dataset (videos/images), the systematization of the accumulated results, the identification of hidden patterns in the biological systems manifested in the response to the effects of damaging factors like ionizing radiation. Moreover, to solve the issues of storing, protection, safety-sharing data and algorithms as tools for analysis amount various information in a convenient framework, the Information System has been developing on the basis of the heterogeneous HybriLIT computing platform (JINR LIT), which has at its disposal both the means for developing such systems and the powerful computing resources of the Govorun. The automatization lets increase the speed and accuracy of complex processing of heterogeneous experimental data. Developing the system allows decreasing a chance of losing any information and having access from any place to continue work. Instead of buying software, we will have constantly technical support and change anything in either algorithms or storage by colleges at MLIT.
Speaker: Ms Inna Kolesnikova -
11:10
→
11:40
The approaches in visualization and quantification of collagen fibers 30m
The extracellular matrix (ECM) is as a very dynamic meshwork of proteins and sugars, dominantly fibrous proteins like collagen, elastin and fibronectin and various specific proteoglycans. ECM has important and multiple roles both in healthy tissues and in the disease. ECM is now recognized as a crucial component of complex cancer microenvironment, and remodeling of ECM follow every step of cancer development, progression and response to treatment [1].
Collagen is the most abundant component of ECM. More than 25 subtypes of collagen have been identified. Among them, collagen type I is the main fibrillar collagen, present in most tissues, forming collagen fibers, responsible for biomechanical properties of tissues [2].
The emerging understanding of collagen organization, remodelling and functions in multiple diseases including cancer could provide new strategies for early cancer detection and cancer therapy.
Methods for visualization and quantification of collagen fibers are expanding.
They vary starting with traditional light microscopy and different histochemical staining including Masson trichrome and picrosirius, immunohistochemistry for different epitopes on collagen fibers, confocal microscopy with fluorescent labelling of collagen and electron microscopy. Other methods such as Raman spectroscopy and atomic force microscopy are being used to examine composition, structure and biomechanical properties of collagen [3, 4]. We will provide a comprehensive review of traditional and novel methods of collagen visualization and quantification with a special reference to nonlinear laser scanning microscopy with second harmonic generation imaging, which is still considered a golden standard in this filed [5, 6, 7].
References
[1] Xu, S., Xu, H., Wang, W. et al. The role of collagen in cancer: from bench to bedside. J Transl Med 2019; 17, 309 https://doi.org/10.1186/s12967-019-2058-1
[2] Sun B. The mechanics of fibrillar collagen extracellular matrix. Cell Rep Phys Sci. 2021; 18;2(8):100515. doi: 10.1016/j.xcrp.2021.100515.
[3] Kular JK, Basu S, Sharma RI. The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J Tissue Eng. 2014; 20;5:2041731414557112. doi: 10.1177/2041731414557112.;
[4] Bhardwaj V, Fabijanic KI, Cohen A, Mao J, Azadegan C, Pittet JC and el. Holistic approach to visualize and quantify collagen organization at macro, micro, and nano-scale. Skin Res and Technology 2022; 28(3):419-426; doi.org/10.1111/srt.13140
[5] Despotović SZ, Milićević ĐN, Krmpot AJ, Pavlović AM, Živanović VD, Krivokapić Z, et al Altered organization of collagen fibers in the uninvolved human colon mucosa 10 cm and 20 cm away from the malignant tumor. Sci Rep. 2020; 14;10(1):6359. doi: 10.1038/s41598-020-63368-y.
[6] Despotović SZ and Ćosić M. The morphological analysis of the collagen fiber straightness in the healthy uninvolved human colon mucosa away from the cancer. 2022; 10: 915644; doi:10.3389/fphy.2022.915644
[7] Chen, X, Nadiarynkh O, Plotnikov S. et al. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat Protoc 2012; 7, 654–669 https://doi.org/10.1038/nprot.2012.009Speaker: Dr Sanja Despotović (Institute of Histology and embryology) -
11:40
→
11:55
Break 15m
-
11:55
→
12:25
Extracting biological meaning from lipidomics data through biostatistics, machine learning and pathway analysis 30m
In recent years, due to significant advancements in mass spectrometry, lipidomics has emerged as a fast growing scientific field that provides deep insights into complex changes in lipidome of human cells and tissues. Human blood is a self-regenerating lipid-rich biological fluid, a tissue that is easily collected in hospitals. Blood/plasma is rich in lipids and related metabolites, and its lipid composition (LIPIDOME) reflects diverse aspects of lipid metabolism and may give us insight into general human physiology in health and disease. Plasma lipidome is a tightly regulated and precisely defined constellation of lipid molecules and disturbances in the plasma lipidome occur in many diseases such as cardiovascular diseases and cancer, but also in conditions that are not directly linked to lipid metabolism. Hence, plasma lipidomics is an emerging tool in an array of clinical diagnostics. The most sought-after goals in the lipidomics community are to identify disease biomarkers, monitor a clinical treatment or confirm a biological hypothesis on a causality between a disease onset/progression and lipid profile. A recommended plasma lipidomics workflow consists of: preanalytics, analytics and post analytics, including study design, research hypotheses, sample collection, demographics data collection, lipid extraction, quality control, liquid chromatography and mass spectrometry, raw data processing, lipid annotation/identification, normalization, lipid quantitation, databases, data sharing and data analysis. The topic of this presentation is data analysis so I will put an accent to it. Lipidomics community nowadays uses dozens of biostatistical tools, artificial intelligence (AI), machine learning (ML) algorithms and chemometrics for data analysis, depending on size of datasets, data structure and the expertise of data scientists involved. Smart analysis of lipidomic data (provided these are of good quality) is of paramount importance for identifying potential biomarkers and understanding disease mechanisms. To accomplish this, they use, besides classical statistics-between groups comparisons, principal component analysis (PCA), orthogonal projections to latent structures discriminant analysis (OPLS-DA), partial least square analysis (PLS) and correlation analysis, all accompanied by various data pattern recognition and data visualization tools. Less often used are ML algorithms such as random forests (RF) and support vector machines (SVM), with only few applying deep learning and neural networks. Finally, to extract biological knowledge, i.e. metabolic pathways affected by a disease or applied treatment, data are subjected to different enrichment analyses, network analyses and pathway analysis. Life scientists working outside clinical setting, such as those investigating neurodegenerative changes in brain will also benefit from applying lipidomics to their model systems.
Speaker: Dr Romana Masnikosa (Vinča Nuclear Research Institute) -
12:25
→
12:50
The Basics of the Radon transform for medical and quantum state tomography 25m
This work provides an accessible introduction to the computational aspects of the Radon transformation, nowadays routinely used to reconstruct the 3D distribution of scatterers from the sequence of their 2D integral projections. The focus will be only on the parallel-beam projections, which directly correlate to a real-world application of X-ray absorption tomography, where measurements of the attenuation of the radiation intensity passing through the specimen at different angles were used to reconstruct a medical tomographic image.
For simplicity, the Radon transform, the inverse Radon transform, and their relationship with the spatial Fourier transform will be introduced by analysis of the 2D medical phantom image of a human head. In this setting, the original grayscale image, represented by a square matrix, can be considered a cross-section through the specimen, with the image’s light intensities representing the density of the absorbers. We shall briefly mention how to accomplish the image reconstruction tasks easily and computationally efficiently using Matlab’s Image Processing Toolbox [1].
In the end, we shall review the use of tomography techniques for reconstructing the quantum state in the phase space using only measurements of the momentum probability distribution done on an ensemble of identically prepared states using homodyne tomography [2] in the case of bosons and quantum beam tomography in the case of fermions [3].
Acknowledgments
The research was funded by the Ministry of Science, Technological Development, and Innovation of the Republic of Serbia (grant No. 451-03-47/2023-01/ 200017)References
[1] The MathWorks, Inc., Image Processing Toolbox, (2023) https://www.mathworks.com/help/images
[2] D. T. Smithey, M. Beck, and M. G. Raymer, Measurement of the Wigner Distribution and the Density Matrix of a Light Mode Using Optical Homodyne Tomography: Application to Squeezed States and the Vacuum, Phys. Rev. Lett. 70, 9 (1993)
[3] S. H. Kienle, M. Freyberger, W .P. Schleich and M. G. Raymer, Quantum Beam Tomography, in Experimental Metaphysics edited by R. S. Cohen, M. Horne, and J. Stachel (Kluwer Academic Publishers, Dordrecht, 1997)Speaker: Dr Marko Ćosić (Vinca institute of nuclear sciencies)
-
10:00
→
10:05
-
-
10:00
→
10:30
Recent studies on nonlinear dynamics of microtubules and DNA 30m
Nonlinear dynamics of two biomolecules is studied. These are a microtubule (MT) and DNA molecule [1,2]. Two mathematical procedures are explained, yielding to three kinds of solitary waves moving through the systems. These waves are modulated solitary waves called breathers, kinks, and bell-type solitons. Regarding DNA, it is shown that a demodulated standing soliton could be crucial for the DNA-RNA transcription [3]. Finally, it is shown that the subsonic waves are stable in MTs, while supersonic ones are not [4].
References
[1] S. Zdravković, Nonlinear Dynamics of DNA Chain, in Nonlinear Dynamics of Nanobiophysics, edited by S. Zdravković and D. Čevizović, Springer, 2022, 29-66.
[2] S. Zdravković, Nonlinear Dynamics of Microtubules, in Nonlinear Dynamics of Nanobiophysics, edited by S. Zdravković and D. Čevizović, Springer, 2022, 263-306.
[3] S. Zdravković, M.V. Satarić, A.Yu. Parkhomenko, and A.N. Bugay, Demodulated standing solitary wave and DNA-RNA transcription, Chaos 28, 113103 (2018)
[4] D. Ranković and S. Zdravković, Two component model of microtubules – subsonic and supersonic solitary waves, Chaos Soliton Fract. 164, 112693 (2022)Speaker: Dr Slobodan Zdravković (Vinča Nuclear Research Institute) -
10:30
→
10:45
System for working with neurocognitive experiments data (MRI/fMRI) on the HybriLIT Heterogeneous Platform 15mSpeaker: Mr Maxim Zuev (MLIT JINR)
-
10:45
→
11:00
Monte Carlo simulation studies of radiation induced damage at cellular and sub-cellular level 15m
In present days, it is of a great interest to further understand the impact of various radiation types on living beings, at cellular and sub-cellular (DNA) level, for the purpose of radiation protection and radiation therapy. Hadronic particles such as protons, carbon ions, and lately helium ions, have various advantages in regards to previous, conventional cancer therapies using photon beams. The number of hadron therapy facilities in the world is rising rapidly, with more than 250,000 patients having received treatment with proton and carbon ion beams. To study the radiobiological effects of ionizing radiation on cells, Monte Carlo simulations that can reproduce and evaluate radiation damage are mandatory. Several track structure Monte Carlo codes exist, however a few of them are openly available to users. The Geant4-DNA software package is publicly available to the user community. The newly released example application molecularDNA, part of Geant4-DNA and publicly available since December 2022, will be presented. This application enables simulations of realistic cancer cell geometries combined with modelling of physical, physico-chemical and chemical stages of cell irradiation, including also radiolythic processes, to give prediction of direct and indirect DNA damage. In the context of future hadron therapу, the benefits of using helium ions will be discussed, as well as other treatment options.
References
[1] K. Chatzipapas, M. Dordevic, S. Zivkovic, N. H. Tran, N. Lampe, D. Sakata, I. Petrovic, A. Ristic-Fira, W.-G. Shin, S. Zein, J. M.C. Brown, I. Kyriakou, D. Emfietzoglou, S. Guatelli, S. Incerti, Geant4 simulation of human cancer cells irradiation with helium ion beams, Phys. Med. Eur. J. Med. Phys. 112, 102613 (2023)
[2] K. Chatzipapas, N. H. Tran, M. Dordevic, S. Zivkovic, S. Zein, W.-G. Shin, D. Sakata, N. Lampe, J. M. Brown, A. Ristic-Fira, I. Petrovic, I. Kyriakou, D. Emfietzoglou, S. Guatelli, S. Incerti, Simulation of DNA damage using Geant4-DNA: an overview of the ”molecularDNA” example application, Precis. Radiat. Oncol., 7, 4 (2023).
[3] D. Sakata, R. Hirayama, W.-G. Shin, M. Belli, M. A. Tabocchini, R. D. Stewart, O. Belov, M. A. Bernal, M.-C. Bordage, J. M.C. Brown, M. Dordevic, D. Emfietzoglou, Z. Francis, S. Guatelli, T. Inaniwa, V. Ivanchenko, M. Karamitros, I. Kyriakou, N. Lampe, Z. Li, S. Meylan, C. Michelet, P. Nieminen, Y. Perrot, I. Petrovic, J. Ramos-Mendez, A. Ristic-Fira, G. Santin, J. Schuemann, H. N. Tran, C. Villagrasa, S. Incerti, Prediction of DNA rejoining kinetics and cell survival after proton irradiation for V79 cells using Geant4-DNA, Phys. Med. 105 102508 (2023).
[4] W.-G. Shin, D. Sakata, N. Lampe, O. Belov, N. H. Tran, I. Petrovic, A. Ristic-Fira, M. Dordevic, M. A. Bernal, M.-C. Bordage, Z. Francis, I. Kyriakou, Y. Perrot, T. Sasaki, C. Villagrasa, S. Guatelli, V. Breton, D. Emfietzoglou, S. Incerti, A Geant4-DNA evaluation of radiation-induced DNA damage on a human fibroblast, Cancers 13, 4940 (2021).Speaker: Dr Miloš Đorđević (Vinča Nuclear Research Institute) -
11:00
→
11:15
Video-based unobtrusive physiological measurements: Perspectives and Opportunities 15mSpeaker: Prof. Nadica Miljkovic (University of Belgrade School of Electrical Engineering)
-
11:15
→
11:30
Break 15m
-
11:30
→
11:45
Quantifying the complexity of the collagen fiber arrangement 15m
It is known that all stages of tumor progression are followed by an extraordinary remodeling of collagen fibers in the extracellular matrix. In the case of colorectal cancer, alignment and straightness have been described. Therefore, a proper measuring of this directional orderliness could be helpful for early detection of tumor presence. An experienced histopathologist's task is to estimate the degree of straightness of collagen fibers. It is a demanding manual endeavor, and possible automatization of the procedure is welcoming. We have gathered images of healthy human colon mucosa, colon mucosa 20 cm away from the cancer, 10 cm away from the cancer, and at the cancer itself via non-linear second harmonic microscopy. In each image, collagen fibers were identified. To quantify the degree of straightness of a collagen bundle in an image, we have divided each fiber into equal-length segments and calculated the distribution of segment angles relative to a fixed, predetermined direction. Information about preferable directions of collagen fibers is contained in the distribution's maxima. The distribution’s mean value and standard deviation are of no use since these values contain no information on the shape of the distribution. We modeled obtained distribution by the polymorphic beta-distribution, a two-parameter family of functions, to study transformation of its extrema. We found that imaged tissue samples could be classified into four categories: patterns without any preferable orientation whose beta-distribution has no extrema, transitional forms whose beta-distribution has a broad single maximum, and highly oriented forms whose beta-distribution has two minima and single very narrow maximum in between. We have demonstrated that variation of collagen directional orderliness can be linked to the shape transformation of the corresponding beta-distributions. We found that samples of healthy individuals have an almost uniform distribution over beta-distribution forms. At 20 cm and 10 cm from the tumor, transitional forms with one broad maximum redistribute over unoriented and highly oriented forms. At the cancer itself, only highly oriented forms are present. To further improve the approach, we have calculated the structural complexities of tissue images. The structure is said to be more complex the more it differs from itself when considered at different scales. We used a measure of structural complexity based on interscale dissimilarities of images. Sample images taken from the healthy tissue, 20 cm and 10 cm away from the tumor and at the tumor itself, were mapped to points in a three-dimensional parametric space. The first two coordinates in this abstract three-dimensional space are beta-distribution parameters, and the third is the corresponding structural complexity value. We have calculated means and covariance matrices of data points for different tissue types. Obtained covariances define ellipsoids in a parametric space. To quantify the distinction between points belonging to different tissue types, we calculated a number of data points within the disjunctive union of each pair of ellipsoids. Consideration of structural complexity enhances the distinction between healthy tissue and tumor by more than 30 percent. We concluded that defined quantifications of directional orderliness and the complexity of collagen fibers could be helpful in the early detection of cancer tissue presence.
References
[1] S. Despotović, and M. Ćosić, “The Morphological Analysis of the Collagen Fiber Straightness in the Healthy Uninvolved Human Colon Mucosa Away From the Cancer”, Frontiers in Physics, vol. 10, 2022,
doi: 10.3389/fphy.2022.915644.
[2] A. A. Bagrov I. A. Iakovlev, A. A. Iliasov, M. I. Katsnelson, and V. V. Mazurenko, “ Multiscale structural complexity of natural patterns”, The Proceedings of the National Academy of Sciences, vol. 117, 2020,
doi: 10.1073/pnas.2004976117.Speaker: Dr Milivoje Hadžijojić (Vinča Nuclear Research Institute) -
11:45
→
12:00
Ultrasound for tissue microscopy 15m
Application of ultrasonic methods for medical diagnostics is widely known due to the delicate effect on living tissues. The methods are less labor-intensiveness compared to X-ray tomography that makes it easier to observe dynamics of biological processes. A distinctive feature of focused ultrasound is the local nature of its interaction with an object. The probing beam has a focal zone with diameter and length determined by the angular aperture and operating frequency of the ultrasound emitter. The frequencies of 1-10 MHz are used in medical diagnostics to observe the structure of human organs. Radiation of higher frequency - 10-200 MHz, is used to study miniature samples, slices and to work with the laboratory animals.
Scanning acoustic microscopy (SAM) provides high-quality images of tissues and cells, comparable to the light microscopy, without any staining and with a short time of scanning. Ultrasonic waves pass through various tissues of the human body with certain speed values. The speed and attenuation of sound waves correlates with elasticity of tissues, thereby indicating their biomechanical properties. Elasticity varies depending on contents, such as: collagen fibers, blood, colloids, mucin, fat, crushed substances, cytoskeleton and so on. Due to the dependence, SAM allows to display changes in the tissues composition. In addition, chemical modifications such as fixation can affect acoustic properties and SAM can tracks these changes over time on the same sample to simplify statistical comparison of the measured data.
SAM was developed in the 70s of the last century at Stanford University [1]. Today, the SAM is modified: the shortest probe pulses and precision mechanical systems for the sensor moving provide a high resolution – lateral and in depth. Wave front of ultrasonic radiation is considered to be flat inside the focal zone of acoustic lens. Due to this, SAM can be used to calculate sound speed and attenuation, tissue thickness, and to image their distribution. The Imaging quality depends on the SAM frequency – the higher frequency corresponds to the smaller wavelength and diameter of focal zone, and a smaller pixel size as a result. Contrast of ultrasonic images depends on the biomechanical properties of tissues. Tissue sections that contain little structural protein possess sound speed and attenuation values that are similar to water in the coupling medium. In contrast, tissue sections containing structural proteins such as collagens have significantly higher the values than water. In general, the resolution of SAM is comparable to the resolution of optical microscopy. But since the principles of contrast formation of these two methods differ, their combination helps to clarify the structural and functional characteristics of tissues and cells as effectively as possible.
In this review, principle of SAM, samples preparation, examples of SAM application will be presented. To illustrate the method potential, the report uses both own original images and the results of other authors published with open access.
References
[1] R. A. Lemons, and C. F. Quate, Acoustic Microscopy, in Physical Acoustics, edited W. P. Mason, and R. N. Thurston, (Academic Press., London, 1979)Speaker: Dr Yulia Petronyuk (N.M. Emanuel Institute of Biochemical Physics RAS) -
12:00
→
12:15
High resolution ultrasound methods in tissue engineering 15m
The fast development of interdisciplinary areas - tissue engineering and regenerative medicine, requires continuous improvement of research approaches and imaging techniques. This report will present the possibilities and perspectives of ultrasound methods for the evaluation of matrixes and tissue engineered constructs (TECs) for regenerative medicine needs. The information obtained by ultrasound includes both the evaluation of TEC morphology (geometry and internal structure of the fabricated matrix, integration into the living organism, settlement with cellular components, formation of extracellular matrix, vascularization and, finally, complete replacement by the organism's own tissues) and elastic properties (measurement of sound velocity, attenuation and spectrum). It should be noted that over the last twenty years a lot of research in this direction has been carried out, but if we delve into the issue in a little more detail, it becomes clear that a more general problem has not been solved: the development of a consistent comprehensive approach to the study of tissue-engineered structures. This situation was due to the fact that acoustic microscopy was used in each individual study to address a specific question on a specific type of matrix, cell, or tissue-engineered construct. Preliminary studies carried out in our laboratory on different types of matrixes before and after implantation in the organism, combined with the undoubted advantages of safety and non-invasiveness, allow us to consider acoustic microscopy as a universal method applicable from the manufacturing stage up to clinical trials, providing clear feedback, will help to develop a general integrated approach.
Speaker: Dr Elena Khramtsova (N.M. Emanuel Institute of Biochemical Physics RAS) -
12:15
→
12:30
Photothermal effect in biomedical applications 15m
When a sample is exposed to EM radiation, part of the excitation energy is absorbed, and part of the absorbed energy is transformed into heat through non-radiative deexcitation-relaxation processes. This effect of optical heating of the sample, so so-called photothermal (PT) effect causes the spread of heat through the sample and through its environment, leading to several phenomena that can be detected in different ways: a change in temperature on the surface of the sample, a change in the spectrum of IR radiation of the sample, bending and/or stretching of the sample, the appearance of a gradient of optical refractive index within the sample (thermal lens effect) or in its environment (mirage effect). All of these phenomena depend on the morphological, thermal, optical, elastic, and other physical properties of the sample that govern the transport processes caused by the effect of an external source of EM radiation. As all these phenomena are non-destructive, the PT effect can be used in in vivo biomedical diagnostics.
The subject of our research is the development of a model for optically induced temperature variations in biological tissues, i.e. solving of direct PT problem. To this end, we take into account the fractal structure of the tissues, thermal memory effects, and blood perfusion effects on the propagation of heat through human tissues. Besides, we analyze the possibilities of tissue image reconstruction and determination of tissue characteristics when some of the PT phenomena dependent on temperature profile are experimentally detected i.e. we solve inverse PT problem. Our results could enable the development of novel non-destructive biomedical diagnostic techniques.
Speaker: Dr Slobodanka Galović (Vinča Nuclear Research Institute) -
12:30
→
13:00
JQuantPro - software for automation of image analysis 30m
Introduction
Fluorescence microscopy techniques are widely used in life science research in general and in particular in radiation biology. These techniques involve labelling of various cellular structures with fluorescent dyes followed by observation and image acquisition using fluorescence wide field and laser scanning microscopy. In radiobiological applications, fluorescence microscopy is often employed to detect and enumerate so-called radiation induced foci (RIF). These foci are submicron fluorescent objects in cell culture and tissue section microscopy images that are associated with formation of radiation-induced DNA damage and response to such damage. Common examples of such RIF are $\gamma$H2AX foci that are formed following phosphorylation of H2AX histone at sites of DNA double strand breaks (DSB) or 53BP1 foci that formed after recruitment of 53BP1 protein, which is an important factor of DNA damage repair, at sites of radiation induced DNA damage. For visualisation of these foci, antibodies labelled with fluorescent dyes are used. Microscope image analysis that include detection, enumeration and investigation of RIF kinetics provide valuable information on the extent and repair of radiation-induced DNA damage and widely used in laboratory [1] and clinical [2] research. Given that visual (manual) image analysis is very time consuming and subjected to operator bias, development of software for automated image analysis presents an important step in improving productivity and objectivity of such image analysis. JQuantPro is the latest version of software that initially was developed for quantitative image analysis aiming at automatic detection, counting and analysing properties of RIF in fluorescence microscopy images [3].Software structure
Apart from performing its major task of detecting and counting RIF, JQuantPro software incorporates a range of additional image and statistical analysis tools that are in part associated with its major task and also can be exploited for solving various general image analysis problems. JQuantPro consists of four modules dedicated for particular stages and types of image analysis. These modules are Image Browser, Object Analysis, Foci Counting and 3D-Counting.One of the main features of JQuantPro is working with lists of images that allows automatic analysis of multiple images. Image Browser module provide tools for selecting images, browsing through the list of selected images and performing a range of general image manipulation operations such as resampling, normalising, equalising, merging colour planes, building projections, cropping, manipulating image display parameters (contrast, brightness) etc.
The major task of detecting and enumeration of RIF requires implementation of at least two jobs. One of these is the identification in images of cell nuclei that are relatively large objects and termed as “objects” in the context of JQuantPro. Another one is the identification and counting of RIF that are relatively small objects within cell nuclei or larger objects. RIF are termed as “foci” in the context of JQuantPro. The first task is implemented in Object Analysis module that performs identification of objects, associated with cell nuclei, generates object collection for each analysed image in the list and saves object collection files for subsequent foci analysis. Apart from preparing object collection files for foci analysis, Object Analysis module include additional quantitative analysis tools for measuring object properties such as object area, intensity, roundness, building distributions and filtering objects.
The second task is implemented in Foci Counting module that starts from importing object collection files generated at previous step by Object Analysis module and overlaying them on images or colour planes (channels) containing foci signal information. The software allows identification and counting of foci in objects from each object collection file for two additional foci images or foci colour channels, for example $\gamma$H2AX and 53BP1 channels labelled with different fluorescent markers. Apart from counting, a range of foci parameters are measured, as well as frequency distribution of nuclei with respect to foci number, and foci co-localisation analysis is performed if two foci channels are involved. Foci Counting module implements an additional tool for creating groups of images from the list, according to experimental groups in biological experiment, with subsequent statistical analysis generating average parameters for each group.
Image analysis algorithms
JQuantPro has been developed aiming to maximise automation in the process of image analysis. The extent of automation depends on the image quality and complexity, and the uniformity of image properties in the list. It ranges from fully automatic identification of objects to manual drawing and editing of each object with a great level of flexibility allowing the operator to interfere with the automatic analysis process.Object identification is based on image segmentation at the first step with classical intensity threshold segmentation as a basic algorithm. This algorithm rarely works alone, as for example for low cell density images of cultured cells, and requires some pre-processing of images for better outcome or post processing corrections. For the pre-processing purpose, JQuantPro implements application of noise filters and Top Hat transformation [4, 5]. Top Hat is a classical topological image transformation that efficiently reveals fine structures in 2D image intensity profile and substantially reduce variable background intensity signal.
In addition to intensity threshold segmentation that segments images based on absolute intensity levels, JQuantPro implements H-Dome transformation based segmentation that relies on detection of local intensity maxima [5], thus assisting in identification of objects with different levels of signal intensity.
Objects are defined in JQuantPro as continuous segmented areas of an image. In the majority of cases, post-processing analysis is required that includes filtering of objects based on their size, shape and intensity parameters and splitting of overlapping objects. The software implements an efficient automatic object splitting algorithm based on the analysis of the object size, shape of its boundary and intensity distribution within the object.
Object identification algorithms are dependent on the range of parameters that software attempts to suggest automatically based, for example, on the analysis of image histogram or the comparison of two images with minimal and maximal number of objects. These parameters include the size of Top Hat structuring element, the height of H-Dome transformation, the minimum and maximum size of the object, the minimum roundness of the object etc. The values of these parameters can be fine-tuned by the operator from operator- controlled analysis of one of the images and then can be applied to the automatic analysis of all other images in the list.
Software validation
JquantPro has been validated by comparing results of automatic and operator manual counting of RIF in a few studies [3,6] Figure 1 demonstrates an example of correlation between manual and automatic counting.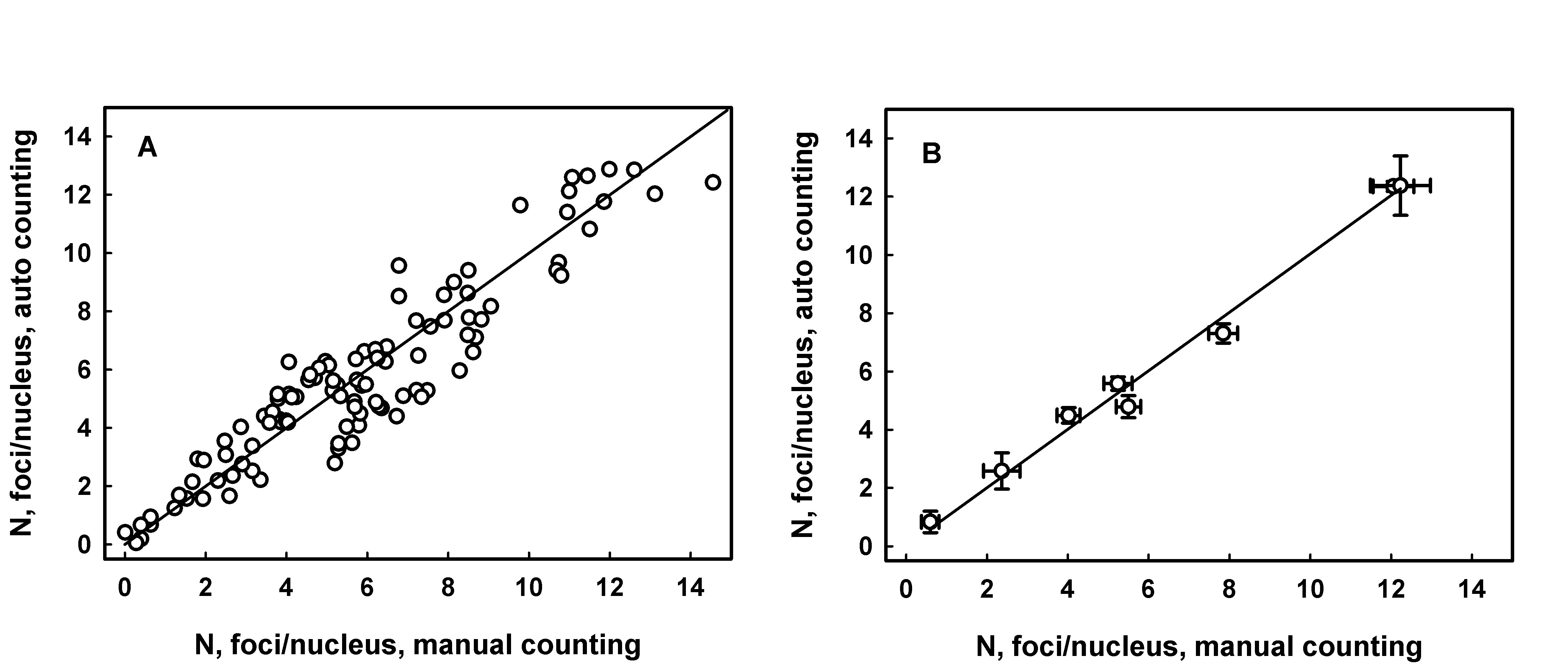
Figure 1. RIF counting analysis in sections of mouse tongue sampled following irradiation of animals with doses from 0.5 to 5 Gy. Panel A: each data point represents average number of foci per nucleus for an individual fluorescence image. The line is generated by linear regression, intercept = 0.26 ± 0.23 (p=0.26, not significant difference from 0), slope = 0.94 ± 0.03, R2 = 0.886. Panel B: each data point represents average number of foci per nucleus for a range of images from an individual experimental group (radiation dose). Error bars indicate the standard error. The line is generated as a result of linear regression analysis, intercept = 0.124 ± 0.308 (p=0.70, not significant difference from 0), slope = 0.99 ± 0.04, R2 = 0.998.
References
[1] P. Lobachevsky, H.B. Forrester, A.Ivashkevich, J. Mason, A.W. Stevenson, C.J. Hall, C.N. Sprung, V.G. Djonov and O.A. Martin. Synchrotron X-Ray Radiation-Induced Bystander Effect: An Impact of the Scattered Radiation, Distance from the Irradiated Site and p53 Cell Status. Frontiers in Oncology, 2021, 11, 685598.
[2] P. Lobachevsky, T. Leong, P. Daly, J, Smith, N. Best, E.R. Thompson, N. Li, I.G. Campbell, R.F. Martin, O.A. Martin. Compromized DNA repair as a basis for identification of cancer radiotherapy patients with extreme radiosensitivity. Cancer Letters, 2016, 383, 212 – 219.
[3] A. N. Ivashkevich, O. A. Martin, A. J. Smith, C. E. Redon, W. M. Bonner, R. F. Martin, P. N. Lobachevsky. γH2AX foci as a measure of DNA damage: a computational approach to automatic analysis. Mutation Research, 711, 49-60, 2011.
[4] M. Van Droogenbroeck, H. Talbot. Fast computation of morphological operations with arbitrary structuring elememnts. Pattern Recognit. Lett., 1996, 17, 1451-1460.
[5] L. Vincent. Morphological grayscale reconstruction in image analysis: applications and efficient algorithms. IEEE Trans. Image Process., 1993, 2 176-201.
[6] L. Jakl, P. Lobachevsky, L. Vokalova, M. Durdik, E. Markova, I. Belyaev. Validation of JCountPro software for efficient assessment of ionizing radiation induced foci in human lymphocytes. International Journal of Radiation Biology, 2016, 92 (12), 766-773.Speaker: Dr Pavel Lobachevsky (Joint Institute for Nuclear Research, Laboratory of Radiation BiIology) -
13:00
→
13:15
The prototype of a web service for a dataset of trajectories of the behavioral test Morris Water Maze 15mSpeaker: Tatevik Bezhanyan (LIT JINR)
-
13:15
→
13:30
The Usage of Complexities for Classification of Neuron Cells 15m
The task of recognizing and classifying neurons is now relevant for histological image analysis. Nowadays, the cells are being counted manually by an expert, and therefore the image processing takes a lot of time. That is why it is important to create a tool to help to classify cells according their mathematical parameters, so that such classification will be consistent with their histological type. Our task was to check the possibility of using the values of cell’s complexity to determine whether it belongs to a histological type. A dataset of histological images of the mouse hippocampus was created for verification. It was marked up using the "cell counter" plugin in ImageJ. This set of images was processed in a special program in Matlab that counts the complexity of the specified areas. According to the obtained data, histograms of the dependence of the number of cells on the value of the complexity they possess were constructed. The results allow us to draw the following conclusions: firstly, the complexity of cells within the same type differ slightly and secondly, the average complexity of cells belonging to different types vary greatly, which means that the value of complexity could be used to differentiate cells by their types. Such a mathematical function could become an important part of a future tool for both pathologists and scientists in this field.
Speaker: Olga Deeva (National Research Nuclear University MEPhI)
-
10:00
→
10:30
-
-
10:00
→
11:30
Tutorial on the use of Python for tasks in Bio-Medical research: Part 1 1h 30m
Service where to copy the code from: HedgeDoc
Connect to server: https://studhub.jinr.ru
Getting started with Jupyter Notebook
- Create directory:
- Rename directory: Right-click on the directory/folder, select “Rename” from the drop-down menu, or select the directory/folder and press “F2”. The directory/folder name must not contain spaces!
- Create Python3 file: Click on “Python3” icon
- Add a new tab (New Launcher):
 Speakers: Anastasia Anikina (Igorevna), Daria Priakhina (JINR, MLIT), Ms Inna Kolesnikova, Margarit Kirakosyan (LIT), Mr Maxim Zuev (MLIT JINR), Oksana Streltsova (JINR), Sara Shadmehri, Tatevik Bezhanyan (LIT JINR)
Speakers: Anastasia Anikina (Igorevna), Daria Priakhina (JINR, MLIT), Ms Inna Kolesnikova, Margarit Kirakosyan (LIT), Mr Maxim Zuev (MLIT JINR), Oksana Streltsova (JINR), Sara Shadmehri, Tatevik Bezhanyan (LIT JINR) -
11:30
→
11:45
Break 15m
-
11:45
→
13:15
Tutorial on the use of Python for tasks in Bio-Medical research: Part 2 1h 30mSpeakers: Anastasia Anikina (Igorevna), Daria Priakhina (JINR, MLIT), Ms Inna Kolesnikova, Margarit Kirakosyan (LIT), Mr Maxim Zuev (MLIT JINR), Oksana Streltsova (JINR), Sara Shadmehri, Tatevik Bezhanyan (LIT JINR)
-
13:15
→
13:30
Closing session 15m
-
10:00
→
11:30
